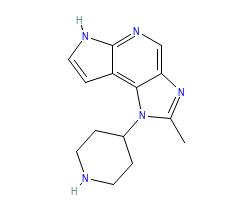
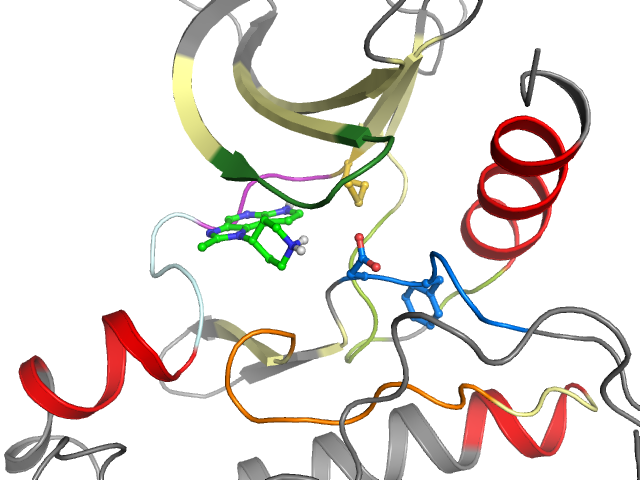
PDB-code
| More entries for 4EHZ | |||||
| 4EHZ | Alternative model: | A | Chain: | A | |
| 4EHZ | Alternative model: | A | Chain: | C | |
| 4EHZ | Alternative model: | A | Chain: | D | |
| 4EHZ | Alternative model: | B | Chain: | A | |
| 4EHZ | Alternative model: | B | Chain: | B | |
| 4EHZ | Alternative model: | B | Chain: | C | |
| 4EHZ | Alternative model: | B | Chain: | D | |
| Structural information | |
| DFG conformation: | in |
| αC-helix conformation: | in |
| G-rich loop angle (distance): | 48.9° (15Å) |
| G-rich loop rotation: | 51.3° |
| Quality Score: | 8 |
| Resolution: | 2.17 Å |
| Missing Residues: | 0 |
| Missing Atoms: | 0 |
| Ligand binding mode | ||||
| Pockets | Subpockets | Waters | ||
| front | FP-I | Cluster I2 I4 I7 I11 | Ligand No No Yes Yes | Protein Yes Yes No Yes |
| Pocket alignment | |
| Uniprot sequence: | RDLGEGHFGKVELVAVKSLDLKKEIEILRNLYENIVKYKGIKLIMEFLPSGSLKEYLPKYLGSRQYVHRDLAARNVLVIGDFGLT |
| Sequence structure: | RDLGEGHFGKVELVAVKSLDLKKEIEILRNLYENIVKYKGIKLIMEFLPSGSLKEYLPKYLGSRQYVHRDLAARNVLVIGDFGLT |
| Ligand affinity | |
| ChEMBL ID: | CHEMBL2159141 |
| Bioaffinities: | 10 records for 2 kinases |
| Species | Kinase (ChEMBL naming) | Median | Min | Max | Type | Records |
|---|---|---|---|---|---|---|
| Homo sapiens | Tyrosine-protein kinase JAK1 | 6.6 | 6.6 | 6.6 | pEC50 | 2 |
| Homo sapiens | Tyrosine-protein kinase JAK1 | 8 | 8 | 8 | pKi | 4 |
| Homo sapiens | Tyrosine-protein kinase JAK2 | 6.7 | 6.7 | 6.7 | pKi | 4 |
Kinase-ligand interaction pattern
• Hydrophobic • Aromatic face-to-face • Aromatic face-to-edge • H-bond donor • H-bond acceptor • Ionic positive • Ionic negative
| I | g.l | II | III | αC | |||||||||||||||
| 1 R 879 | 2 D 880 | 3 L 881 | 4 G 882 | 5 E 883 | 6 G 884 | 7 H 885 | 8 F 886 | 9 G 887 | 10 K 888 | 11 V 889 | 12 E 890 | 13 L 891 | 14 V 905 | 15 A 906 | 16 V 907 | 17 K 908 | 18 S 909 | 19 L 910 | 20 D 921 |
| • | • | • | • | ||||||||||||||||
| αC | b.l | IV | |||||||||||||||||
| 21 L 922 | 22 K 923 | 23 K 924 | 24 E 925 | 25 I 926 | 26 E 927 | 27 I 928 | 28 L 929 | 29 R 930 | 30 N 931 | 31 L 932 | 32 Y 933 | 33 E 935 | 34 N 936 | 35 I 937 | 36 V 938 | 37 K 939 | 38 Y 940 | 39 K 941 | 40 G 942 |
| • | |||||||||||||||||||
| IV | V | GK | hinge | linker | αD | αE | |||||||||||||
| 41 I 943 | 42 K 953 | 43 L 954 | 44 I 955 | 45 M 956 | 46 E 957 | 47 F 958 | 48 L 959 | 49 P 960 | 50 S 961 | 51 G 962 | 52 S 963 | 53 L 964 | 54 K 965 | 55 E 966 | 56 Y 967 | 57 L 968 | 58 P 969 | 59 K 970 | 60 Y 993 |
| • | • | •• | •• | • | • | • | |||||||||||||
| αE | VI | c.l | VII | VIII | x | ||||||||||||||
| 61 L 994 | 62 G 995 | 63 S 996 | 64 R 997 | 65 Q 998 | 66 Y 999 | 67 V 1000 | 68 H 1001 | 69 R 1002 | 70 D 1003 | 71 L 1004 | 72 A 1005 | 73 A 1006 | 74 R 1007 | 75 N 1008 | 76 V 1009 | 77 L 1010 | 78 V 1011 | 79 I 1019 | 80 G 1020 |
| • | • | • | |||||||||||||||||
| DFG | a.l | ||||||||||||||||||
| 81 D 1021 | 82 F 1022 | 83 G 1023 | 84 L 1024 | 85 T 1025 | |||||||||||||||
| •• | |||||||||||||||||||
Interaction pattern search
Search KLIFS for kinase-ligand complexes with similar interaction patterns: