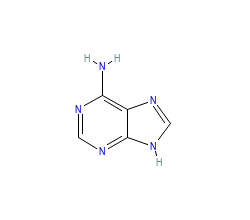
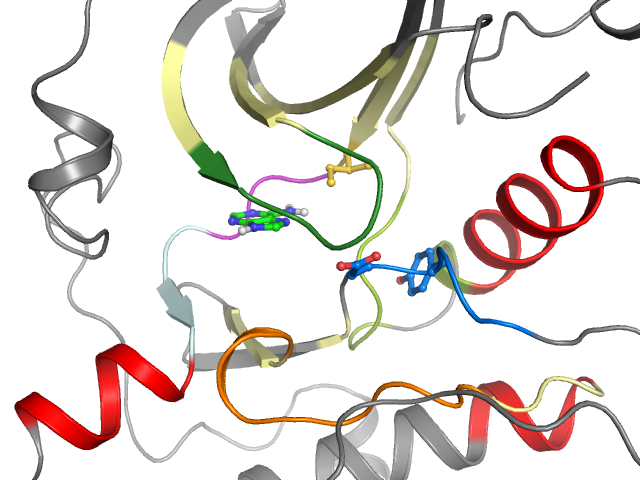
PDB-code
| More entries for 4DC2 | |||||
| 4DC2 | Alternative model: | A | Chain: | A | |
| Structural information | |
| DFG conformation: | in |
| αC-helix conformation: | in |
| G-rich loop angle (distance): | 46.8° (13.9Å) |
| G-rich loop rotation: | 66° |
| Quality Score: | 8.6 |
| Resolution: | 2.4 Å |
| Missing Residues: | 0 |
| Missing Atoms: | 14 |
| Ligand binding mode | ||||
| Pockets | Subpockets | |||
| front | ||||
| Pocket alignment | |
| Uniprot sequence: | RVIGRGSYAKVLLYAMKVVWVQTEKHVFEQASPFLVGLHSCFFVIEYVNGGDLMFHMQRYLHERGIIYRDLKLDNVLLLTDYGMC |
| Sequence structure: | RVIGRGSYAKVLLYAMRVVWVQTEKHVFEQASPFLVGLHSCFFVIEYVNGGDLMFHMQRYLHERGIIYRDLKLDNVLLLTDYGMC |
| Ligand affinity | |
| ChEMBL ID: | CHEMBL226345 |
| Bioaffinities: | No (p)Ki/(p)IC50/(p)EC50 values for kinases found (confidence ≥ 8). |
Kinase-ligand interaction pattern
• Hydrophobic • Aromatic face-to-face • Aromatic face-to-edge • H-bond donor • H-bond acceptor • Ionic positive • Ionic negative
| I | g.l | II | III | αC | |||||||||||||||
| 1 R 248 | 2 V 249 | 3 I 250 | 4 G 251 | 5 R 252 | 6 G 253 | 7 S 254 | 8 Y 255 | 9 A 256 | 10 K 257 | 11 V 258 | 12 L 259 | 13 L 260 | 14 Y 270 | 15 A 271 | 16 M 272 | 17 R 273 | 18 V 274 | 19 V 275 | 20 W 288 |
| • | • | • | |||||||||||||||||
| αC | b.l | IV | |||||||||||||||||
| 21 V 289 | 22 Q 290 | 23 T 291 | 24 E 292 | 25 K 293 | 26 H 294 | 27 V 295 | 28 F 296 | 29 E 297 | 30 Q 298 | 31 A 299 | 32 S 300 | 33 P 303 | 34 F 304 | 35 L 305 | 36 V 306 | 37 G 307 | 38 L 308 | 39 H 309 | 40 S 310 |
| IV | V | GK | hinge | linker | αD | αE | |||||||||||||
| 41 C 311 | 42 F 319 | 43 F 320 | 44 V 321 | 45 I 322 | 46 E 323 | 47 Y 324 | 48 V 325 | 49 N 326 | 50 G 327 | 51 G 328 | 52 D 329 | 53 L 330 | 54 M 331 | 55 F 332 | 56 H 333 | 57 M 334 | 58 Q 335 | 59 R 336 | 60 Y 358 |
| • | • | • | •• | ||||||||||||||||
| αE | VI | c.l | VII | VIII | x | ||||||||||||||
| 61 L 359 | 62 H 360 | 63 E 361 | 64 R 362 | 65 G 363 | 66 I 364 | 67 I 365 | 68 Y 366 | 69 R 367 | 70 D 368 | 71 L 369 | 72 K 370 | 73 L 371 | 74 D 372 | 75 N 373 | 76 V 374 | 77 L 375 | 78 L 376 | 79 L 384 | 80 T 385 |
| • | •• | ||||||||||||||||||
| DFG | a.l | ||||||||||||||||||
| 81 D 386 | 82 Y 387 | 83 G 388 | 84 M 389 | 85 C 390 | |||||||||||||||
Interaction pattern search
Search KLIFS for kinase-ligand complexes with similar interaction patterns: