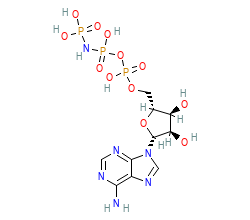
PDB-code
| More entries for 1J1B | |||
| 1J1B | Chain: | A | |
| Structural information | |
| DFG conformation: | in |
| αC-helix conformation: | in |
| G-rich loop angle (distance): | 52.7° (16.2Å) |
| G-rich loop rotation: | 60.5° |
| Quality Score: | 8 |
| Resolution: | 1.8 Å |
| Missing Residues: | 0 |
| Missing Atoms: | 0 |
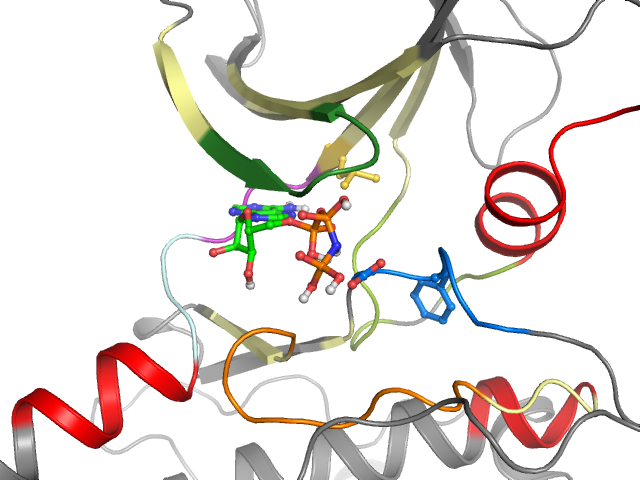
| Ligand binding mode | ||||
| Pockets | Subpockets | Waters | ||
| front | Cluster I1 I4 I5 I7 I11 | Ligand No No Yes Yes Yes | Protein Yes Yes Yes No No | |
| Pocket alignment | |
| Uniprot sequence: | KVIGNGSFGVVYQVAIKKVFKNRELQIMRKLDCNIVRLRYFNLVLDYVP-ETVYRVARHYIHSFGICHRDIKPQNLLLLCDFGSA |
| Sequence structure: | KVIGNGSFGVVYQVAIKKVFKNRELQIMRKLDCNIVRLRYFNLVLDYVP_ETVYRVARHYIHSFGICHRDIKPQNLLLLCDFGSA |
| Ligand affinity | |
| ChEMBL ID: | CHEMBL1230989 |
| Bioaffinities: | No (p)Ki/(p)IC50/(p)EC50 values for kinases found (confidence ≥ 8). |
Kinase-ligand interaction pattern
• Hydrophobic • Aromatic face-to-face • Aromatic face-to-edge • H-bond donor • H-bond acceptor • Ionic positive • Ionic negative
| I | g.l | II | III | αC | |||||||||||||||
| 1 K 560 | 2 V 561 | 3 I 562 | 4 G 563 | 5 N 564 | 6 G 565 | 7 S 566 | 8 F 567 | 9 G 568 | 10 V 569 | 11 V 570 | 12 Y 571 | 13 Q 572 | 14 V 582 | 15 A 583 | 16 I 584 | 17 K 585 | 18 K 586 | 19 V 587 | 20 F 593 |
| • | • | • | • | • | |||||||||||||||
| αC | b.l | IV | |||||||||||||||||
| 21 K 594 | 22 N 595 | 23 R 596 | 24 E 597 | 25 L 598 | 26 Q 599 | 27 I 600 | 28 M 601 | 29 R 602 | 30 K 603 | 31 L 604 | 32 D 605 | 33 C 607 | 34 N 608 | 35 I 609 | 36 V 610 | 37 R 611 | 38 L 612 | 39 R 613 | 40 Y 614 |
| IV | V | GK | hinge | linker | αD | αE | |||||||||||||
| 41 F 615 | 42 N 629 | 43 L 630 | 44 V 631 | 45 L 632 | 46 D 633 | 47 Y 634 | 48 V 635 | 49 P 636 | 50 _ _ | 51 E 637 | 52 T 638 | 53 V 639 | 54 Y 640 | 55 R 641 | 56 V 642 | 57 A 643 | 58 R 644 | 59 H 645 | 60 Y 671 |
| • | •• | •• | • | ||||||||||||||||
| αE | VI | c.l | VII | VIII | x | ||||||||||||||
| 61 I 672 | 62 H 673 | 63 S 674 | 64 F 675 | 65 G 676 | 66 I 677 | 67 C 678 | 68 H 679 | 69 R 680 | 70 D 681 | 71 I 682 | 72 K 683 | 73 P 684 | 74 Q 685 | 75 N 686 | 76 L 687 | 77 L 688 | 78 L 689 | 79 L 698 | 80 C 699 |
| • | • | • | |||||||||||||||||
| DFG | a.l | ||||||||||||||||||
| 81 D 700 | 82 F 701 | 83 G 702 | 84 S 703 | 85 A 704 | |||||||||||||||
| • | |||||||||||||||||||
Interaction pattern search
Search KLIFS for kinase-ligand complexes with similar interaction patterns: