PDB-code
| More entries for 7T4T | |||||
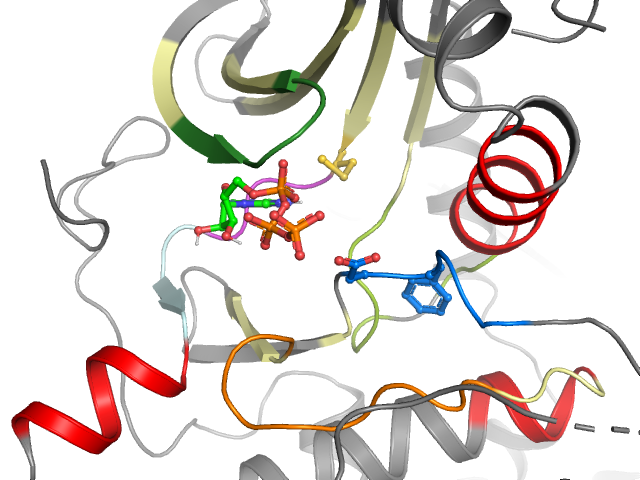
| 7T4T | Alternative model: | B | Chain: | A | |
| Structural information | |
| DFG conformation: | in |
| αC-helix conformation: | in |
| G-rich loop angle (distance): | 59.8° (17.9Å) |
| G-rich loop rotation: | 56.4° |
| Quality Score: | 8 |
| Resolution: | 2.08 Å |
| Missing Residues: | 0 |
| Missing Atoms: | 0 |
| Ligand binding mode | ||||
| Pockets | Subpockets | |||
| front | FP-II | |||
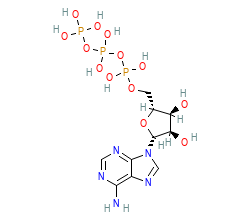
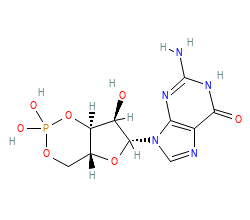
| Allosteric Ligand | |||||||||||||||||||
| Allosteric ligand: | PCG | ||||||||||||||||||
 | |||||||||||||||||||
| Pocket alignment | |
| Uniprot sequence: | DTLGVGGFGRVELFAMKILHIRSEKQIMQGAHDFIVRLYRTYMLMEACLGGELWTILRDYLHSKGIIYRDLKPENLILLVDFGFA |
| Sequence structure: | DTLGVGGFGRVELFAMKILHIRSEKQIMQGAHDFIVRLYRTYMLMEACLGGELWTILRDYLHSKGIIYRDLKPENLILLVDFGFA |
| Ligand affinity | |
| ChEMBL ID: | CHEMBL14249 |
| Bioaffinities: | 4 records for 2 kinases |
| Species | Kinase (ChEMBL naming) | Median | Min | Max | Type | Records |
|---|---|---|---|---|---|---|
| Homo sapiens | MAP kinase-activated protein kinase 2 | 5.2 | 4.9 | 5.6 | pIC50 | 3 |
| Homo sapiens | Mitogen-activated protein kinase kinase kinase 7 | 5.6 | 5.6 | 5.6 | pKd | 1 |
Kinase-ligand interaction pattern
• Hydrophobic • Aromatic face-to-face • Aromatic face-to-edge • H-bond donor • H-bond acceptor • Ionic positive • Ionic negative
| I | g.l | II | III | αC | |||||||||||||||
| 1 D 379 | 2 T 380 | 3 L 381 | 4 G 382 | 5 V 383 | 6 G 384 | 7 G 385 | 8 F 386 | 9 G 387 | 10 R 388 | 11 V 389 | 12 E 390 | 13 L 391 | 14 F 402 | 15 A 403 | 16 M 404 | 17 K 405 | 18 I 406 | 19 L 407 | 20 H 420 |
| • | • | • | • | • | • | • | • | ||||||||||||
| αC | b.l | IV | |||||||||||||||||
| 21 I 421 | 22 R 422 | 23 S 423 | 24 E 424 | 25 K 425 | 26 Q 426 | 27 I 427 | 28 M 428 | 29 Q 429 | 30 G 430 | 31 A 431 | 32 H 432 | 33 D 434 | 34 F 435 | 35 I 436 | 36 V 437 | 37 R 438 | 38 L 439 | 39 Y 440 | 40 R 441 |
| IV | V | GK | hinge | linker | αD | αE | |||||||||||||
| 41 T 442 | 42 Y 450 | 43 M 451 | 44 L 452 | 45 M 453 | 46 E 454 | 47 A 455 | 48 C 456 | 49 L 457 | 50 G 458 | 51 G 459 | 52 E 460 | 53 L 461 | 54 W 462 | 55 T 463 | 56 I 464 | 57 L 465 | 58 R 466 | 59 D 467 | 60 Y 489 |
| • | • | •• | •• | ||||||||||||||||
| αE | VI | c.l | VII | VIII | x | ||||||||||||||
| 61 L 490 | 62 H 491 | 63 S 492 | 64 K 493 | 65 G 494 | 66 I 495 | 67 I 496 | 68 Y 497 | 69 R 498 | 70 D 499 | 71 L 500 | 72 K 501 | 73 P 502 | 74 E 503 | 75 N 504 | 76 L 505 | 77 I 506 | 78 L 507 | 79 L 515 | 80 V 516 |
| • | • | ||||||||||||||||||
| DFG | a.l | ||||||||||||||||||
| 81 D 517 | 82 F 518 | 83 G 519 | 84 F 520 | 85 A 521 | |||||||||||||||
Interaction pattern search
Search KLIFS for kinase-ligand complexes with similar interaction patterns: